Mapping of Peptides
Introduction
Recent advances in high-throughput techniques like phage display resulted in an enormous amount of PDZ-mediated interaction data. But the most often these techniques generate the interactions involving random peptides. To make use of these data, it is important to map random peptides to human C-terminal proteome since PDZ domains recognize C-terminal peptide.
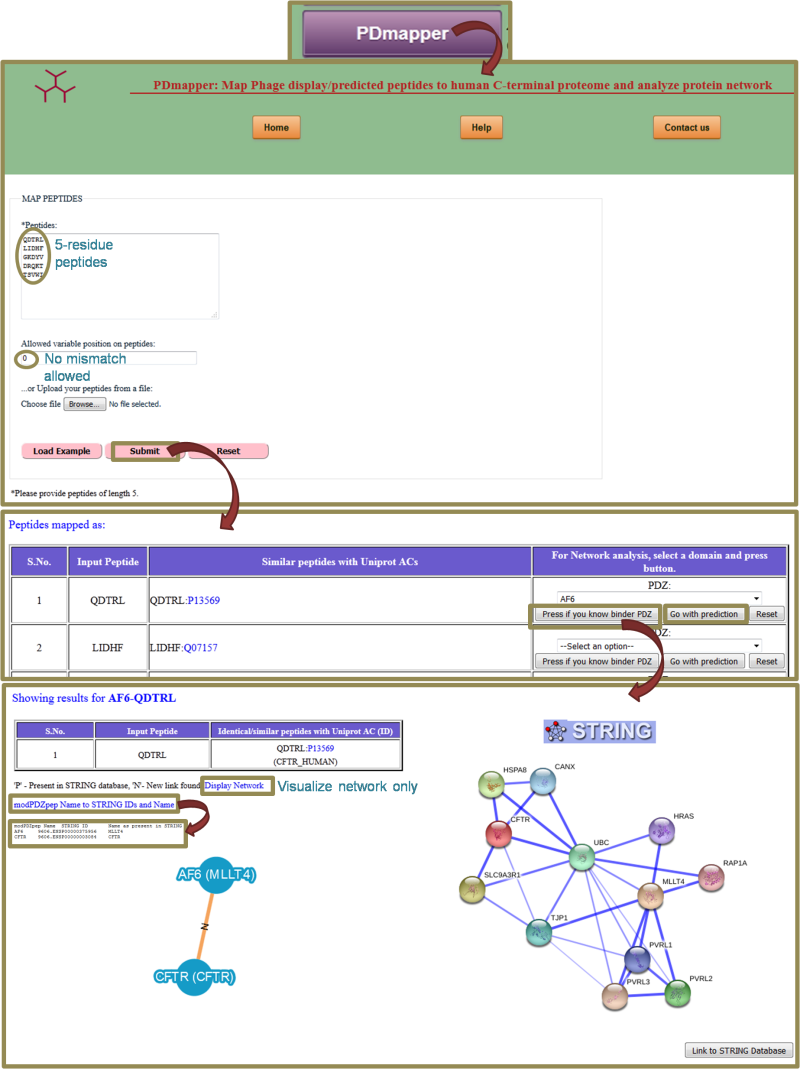
UsagePDmapper takes as an input last five residues of phage display peptide and provide its Uniprot AC. It also allows to have some variability in the peptide sequence to match Uniprot AC but it sticks to the criteria of having hydrophobic C-terminal residue required for interaction to the PDZ domains. Since not all the phage display peptides show exact match to C-terminal proteome, it provides added advantage to pick up for proteins with similar C-terminus. One can also generate the PDZ-protein network with these similar peptides by selecting the domain from the option list and clicking Go. It has two options- if you already have information about binder PDZ domain, you can go for first option or for second option, in which predicted binding BT-score is also provided. We suggest a more stringent cutoff of -3 to classify binder and non-binder peptides based on our analysis. Lower the score, better the binding ability. The reliability of network can be assessed by comparing the developed network with STRING database interactions provided on the page itself. Here, edges are labeled with either 'P' or 'N' meaning already present in STRING or a new link has been found. This new link may be a direct link between two proteins which are interacting through some other protein in the STRING. 'Display Network' gives a larger view of the network. Link 'modPDZpep Name to STRING IDs and name' provides the protein names as used in modPDZpep and STRING database with their STRING IDs.
 |